
Tuesday, September 30, 2014
Electrophoresis V.S Western Blotting for detection of a protein
So after some intense dialog with the Biker Scout and C-3PO I think that I may be a bit closer to a project idea, even though it seems very far away from my goal upon first glance. This semester I will be using electrophoresis to isolate a specific protein. I will then use western blotting to introduce antibodies to the protein. Once I become fluent in this process my hope is to actually go after more specific human proteins/antibodies. I am curious to know if the antibodies produced from the beta amyloid protein could be used to create a test that would show the varying degree of beta amyloid proteins if they were present in an individual's blood. The first question that I had was; "Can beta-amyloid proteins be detected/isolated from urine and/or blood"? The answer is yes, they can be detected in blood, cerebral spinal fluid, and urine. My next question was,"Why is there not an inexpensive urinalysis (the least invasive procedure for the patient) that could detect the amount of beta amyloid proteins excreted through urine"? If the theory of the accumulation of these proteins or plaques is what causes the cognitive decline associated in Alzheimer's Disease is correct, than why are we not incorporating a device that will detect these plaques BEFORE they have time to build up? Many health care professionals already incorporate urinalysis into their annual physical protocol, why are we not monitoring the production of these proteins? Currently, the testing for these proteins is extremely costly and is almost always done on individuals that are ALREADY experiencing cognitive decline. I guess what I am asking is; when will we start to become proactive and get ahead of this disease, instead of ignoring it until it takes up space in our heads?


Thursday, September 25, 2014
Change in Plan
So apparently FISHing is super expensive, so I will not be doing it this semester. So with a heavy heart I will have to choose another project. I have always been curious about the beta amyloid plaques that accumulate in the brains of individuals affected with Alzheimer's Disease. These plaques are actually a sequence of 55 amino acids that most every person produces. The ability to excrete these proteins through urine reduces the probability of one being affected by this disease. When a person's body does not excrete the beta amyloid proteins they start to build up, like plaque on teeth. The once happy neurons frolicking happily in the brain are now plagued by these protein formations that prevent communication between one another. It takes many years for these formations to effect cognition which is why Alzheimer disease is usually reserved for the elderly population. I would like to see if I can isolate protein from blood in order to eventually acquire an antibody that I might study. If this project is successful, I would like to try to isolate beta amyloid protein from human blood. I wonder if I could start this process with blood from another organism?
Wednesday, September 17, 2014
What is the Difference Between FISH and a Karyotype?
FISH analysis will tell you if a specific chromosome is present and how many chromosomes there are and is much faster than a karyotype. FISH is used commonly for diagnosing down syndrome during pregnancy using amniotic fluid. Downs syndrome is diagnosed when there is a trisonomy on chromosome #21 (that is a number sign not a hash-tag). Now, what it will not tell you is the structural composition of the chromosome (like a karyotype) which is equally as important as knowing how many chromosomes are present. FISH analysis can take from 7-10 days. A karyotype can take up to two weeks to yield results. So, they are similar but very different procedures. Comparing FISHing to Karyotyping (not sure if either of those words are real words but you understand me for all intents and purposes) is like comparing an X-ray to an MRI. The X-ray is great for confirming the general location of pain that a patient is experiencing, but an MRI may be needed if there are further questions regarding structural integrity.So this semester I will learn to FISH, and if it is fun maybe I will try to karyotype (not myself though because I do not need to know that much about myself. It is already hard enough to compartmentalize the information I already have) totally kidding (kind of) :)
 |
| FISH (property of Breen lab at NCSU) |
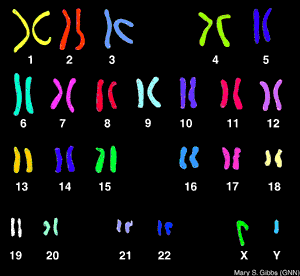 | ||
| KARYOTYPE |
Tuesday, September 16, 2014
FISHing
Hi there, this is my first ever blog so please keep that in mind while reading. I guess I will start with the first two questions that I had when I learned about FISH; what is FISH and why do I care? Fluorescence in situ hybridization (FISH) is a laboratory technique for detecting and locating a specific DNA sequence on a chromosome. The technique relies on exposing chromosomes to a small DNA sequence called a probe that has a fluorescent molecule attached to it. The probe sequence binds to its corresponding sequence on the chromosome.(http://ghr.nlm.nih.gov/glossar y=fluorescentinsituhyb ridization). FISH is used by geneticists in order to diagnose abnormalities within chomosomes, which can save lives. That is why I care. I am a little nervous about attempting this project because it takes extreme precision and requires that measurements be conducted at specific times. I am super excited and ready to get started!

© 2005 Nature Publishing Group Speicher, M. R. et al. The new cytogenetics: blurring the boundaries with molecular biology. Nature Reviews Genetics 6, 784 (2005). All rights reserved.
Subscribe to:
Posts (Atom)
 Figure 1: Principles of fluorescence in situ hybridization (FISH).
Figure 1: Principles of fluorescence in situ hybridization (FISH).